A Primer on DNA Synthesis
Surprise surprise, we have a guest writer!
At BioCompute, we have always placed a lot of emphasis on community. That’s why I started the community for young biotech entrepreneurs in April last year, and recently launched the community for scientists and engineers in the US looking to come back to India. These communities have helped us on rough days (including procurement and hiring), and given me a front row seat on diverse perspectives.
That’s why we are now expanding our blog to bring in folks who want to break down complex DNA and data storage concepts for the non-expert. This is meant to be an extension of some of the research heavy pieces that I have written for the blog earlier (you can check them out here, here and here).
I started thinking of this idea when I first spoke to Saraswathi, a masters student in biotechnology at the Indian Institute of Technology (IIT) Guwahati, in June this year. She asked me whether we are open to having folks from outside BioCompute write for our blog, and back then I was in doubt. This is something that I started almost as a personal blog, and grew into being a company blog. I was unsure whether folks from outside BioCompute would be able to bring the spirit of our work into the world with as much care as we do.
But Saraswathi kept at it relentlessly, writing multiple follow up emails to me, and offering to put together a draft essay without any guarantee that it would see the light of the day. And when I read her work, I knew this had to come into the world. So we worked together on editing it, and after 4ish months of back and forth, her piece on DNA synthesis (how it works with some illustrations) is here for you to read.
*******
Some definitions:
Nucleotide: Refers to A, T, G or C - fundamental units that the molecule DNA is made of
Codon: Refers to a set of three nucleotides in any order
The Building Blocks
Nucleotides are like Legos: they are building blocks used to build something of the creator’s fancy. Legos build castles, nucleotides build the very code that makes us all what we are—DNA. The entire blueprint for our existence is stored in these stretches of A, T, G, and C. So why can’t we try to store all the data we hoard and multiply, in the same format?
As the world relentlessly flies through this digital era, isn’t the most important thing accessible storage for all our data? DNA data storage is a booming industry owing to its immense potential in reducing the need for more silicon hardware. But one of the major struggles we face is the cost and chemical load that is brought in by the synthesis process of DNA.
how is DNA synthesized? The traditional methods involve the addition of free nucleotides to a growing sequence one at a time. This is done using 2 methods:
(a) chemical synthesis and (b) enzymatic synthesis.
Chemical Synthesis
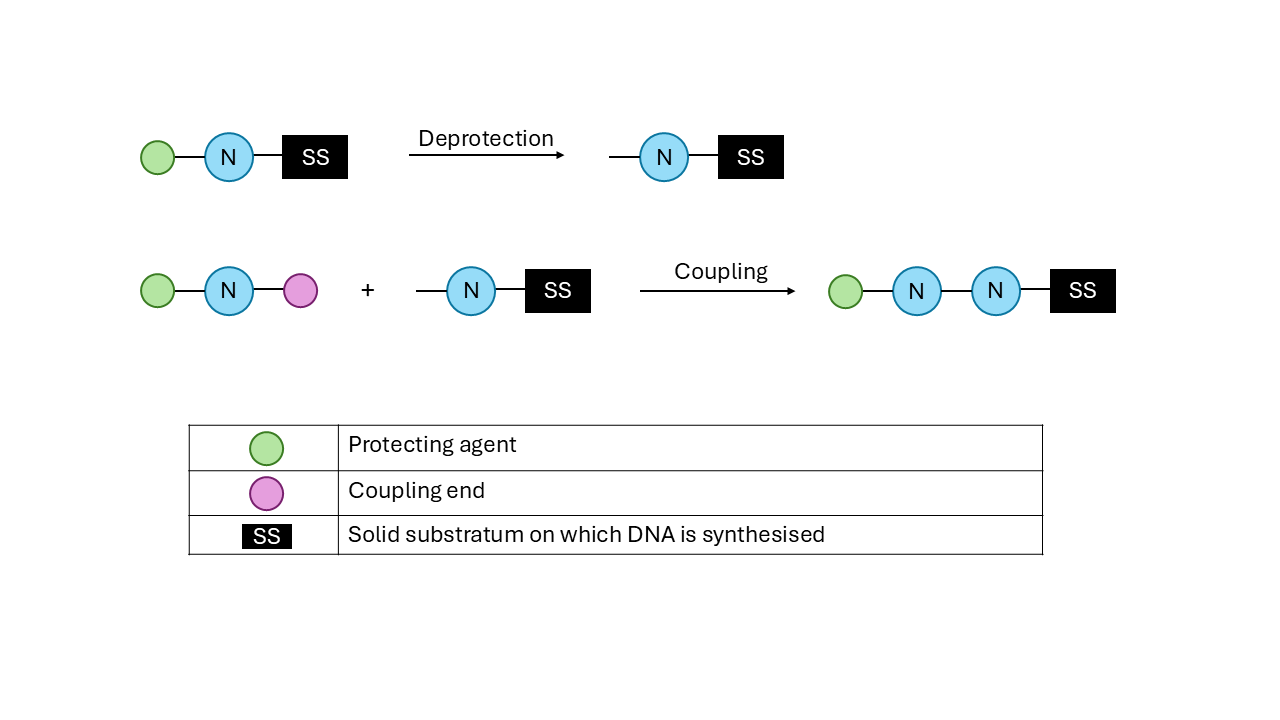
In chemical synthesis, we have nucleotides that have one free end and one protected end. This is done to avoid random reactions between nucleotides. This protecting agent is removed at each step to allow the next nucleotide to attach to it. This is a highly accurate method owing to this protection-deprotection cycle.
Check out the image below for context. And here is an explanatory video.
Image Description: Schematic for chemical synthesis of DNA. Figure by author.
Chemical synthesis is a highly accurate method owing to this protection-deprotection cycle. Despite this, it has its limitations. It employs harsh chemicals for the deprotection steps, which can be harmful to both the handler and the environment in the long term. Purifying the sequences becomes cumbersome and costly as the sequences get longer. That’s why enzymatic synthesis becomes a compelling alternative.
Enzymatic Synthesis
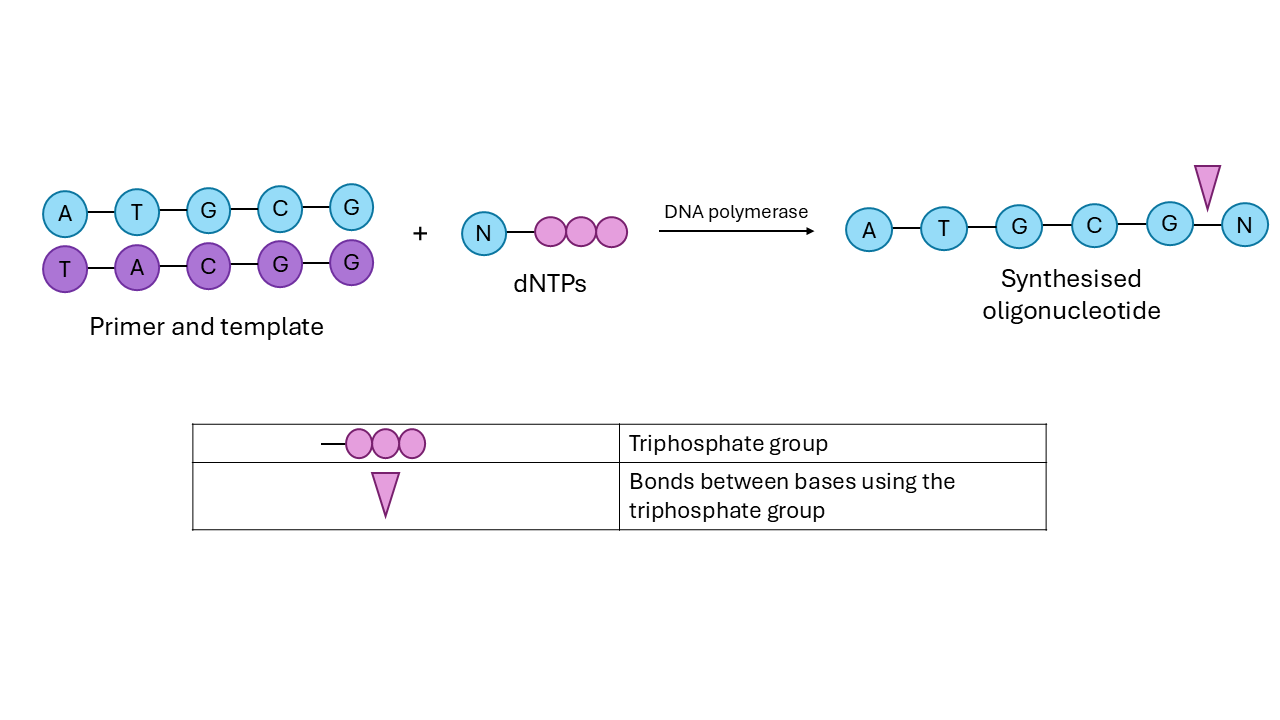
Enzymatic synthesis uses a template for the nucleotides to follow. Nucleotides bases have complementary pairs: A to T, and G to C. The template will be complementary to our desired sequence. A sequence of bases called a primer will initiate this sequence formation. We also add free nucleotides with a triphosphate group attached to one end (they are called deoxyribonucleotide triphosphates, or dNTPs for short). Enzymes called ‘DNA polymerases’ will be used to facilitate the addition of the dNTPs at every step.
Shared below is a schematic for the process. If you want to dive deeper, here is a series of videos on this process.
Image Description: Schematic for enzymatic synthesis of DNA. Figure by author.
Although the chemicals used in enzymatic synthesis are less harmful, it is more error-prone than the chemical method, as we provide all the nucleotides at once, and allow the additions to happen altogether within the reaction mix.
Best of Both Worlds
A common drawback of both chemical and enzymatic synthesis methods is that we cannot easily modify or tag the DNA sequences. Tagging or modifying bases is a useful tool for storing information in DNA. But doing so might affect its coupling efficiency to the adjacent base, rendering it useless. This begs the question: is there a better method of DNA synthesis that can leverage the benefits of existing methods while mitigating their limitations?
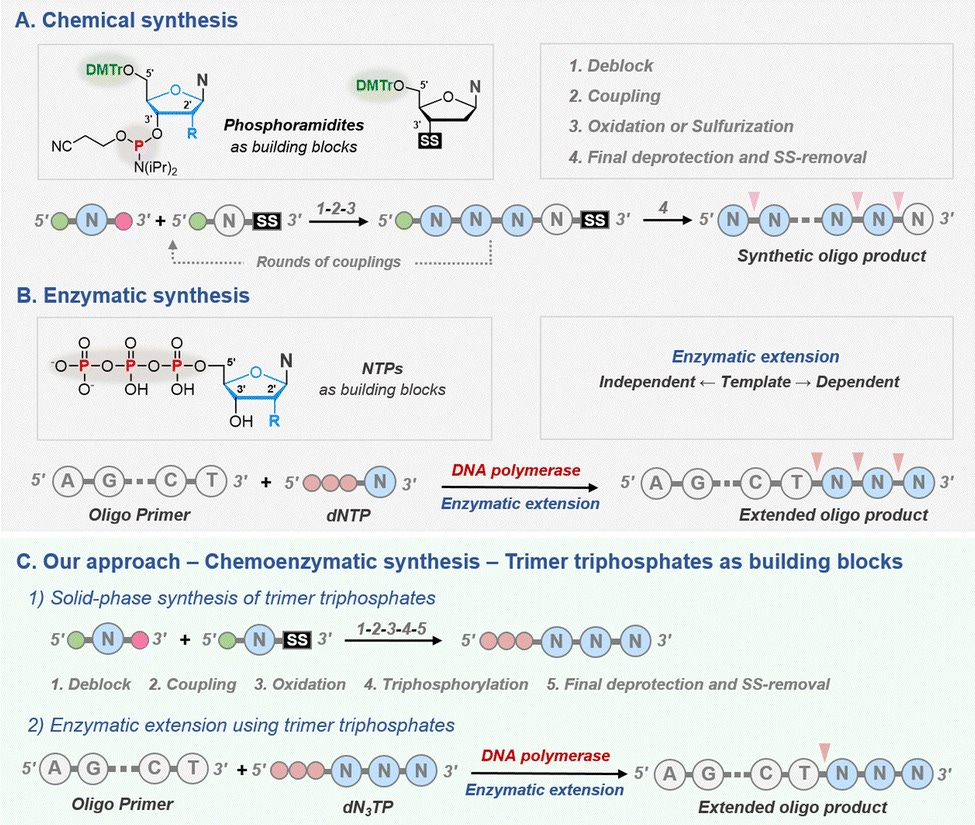
A study published by a team of French and Swiss researchers in 2023 brings a new perspective to DNA synthesis. They build trinucleotide blocks and then arranged them one after another to build the whole sequence, instead of building the sequence one nucleotide at a time. But what is the benefit of doing it this way?
They get the best of both worlds: the fidelity of chemical synthesis and the ease of enzymatic synthesis. They used a trinucleotide, dT3TP (deoxy tri-thymine triphosphate), as the model substrate (as opposed to the single free nucleotide used in the traditional methods). These trimers were created using the chemical synthesis method (smaller sequences ensure lower use of harsh chemicals and easier purification) and extension using these dT3TPs were done using enzymatic methods.
Image Description: Research workflow proposed by Sabat et al. 2023
They used two templates to study the activity of these trimers. Template 1 was a test template with a 15 nucleotide (abbreviated as nt) primer region followed by a series of A’s called a polyA tail (complementary to T, which is what our model is made of). This polyA tail had the space for 4 codons (total of 27 nt). Template 2 had the same 15 nt primer followed by a longer tail sequence at the end. This tail had stretches of 3 A’s (represented as dA3) sprinkled in the middle. This was a total of 47 nt. Once a successful extension was seen in the test template, they would confirm it with the second template.
When they tested their substrate with the test DNA template, the trimers were seen to not extend the primer at all, and even showed degradation. This made the researchers wonder whether the phosphodiester bonds, those which connect the nucleotides together to form the backbone of a DNA sequence, had something to do with stability. They then came up with a new model, which had phosphorothioate bonds ( the P=O structure in the phosphodiester bond is replaced with P=S for better stability against degradation). This modified trimer extended the primer completely for the test template as well as the dA3 interspaced template, proving that using codons as building blocks for oligonucleotide synthesis is possible.
What does this mean for the future of DNA data storage?
This method improves the fidelity of DNA synthesis while reducing its harmful effects on the environment as well as its handlers. It also provides an extra benefit by allowing us to modify and tag the internal bases easily without affecting coupling efficiency of the trimer: the flanking bases (i.e. bases at the end of a DNA sequence) remain unaffected.
This method reduces the time taken as well as the money spent on synthesizing DNA, which can be beneficial while designing the sequence to store data in. Reduced effort and cost can make DNA data storage more accessible and affordable to everyone.
******
Meet the Author:
I’m Saraswathi - a bench scientist by day and a writer by night. I’m currently pursuing my masters in biotechnology at IIT Guwahati, and previously did an undergrad at SASTRA University in Thanjavur. I am an aspiring science communicator. You can find my blog ‘Science Simplified’ on Medium (yes, Anagha tried to convince me to shift to Substack, but that’s for another time).
Apart from that, I love singing and listening to music (Taylor Swift tops my Spotify every single year). I also enjoy crocheting and watching sitcoms. Catch me on a random day and I’m probably scrolling through Instagram or watching The Office. I’m also a bit of a podcast nut, so if you ever need recommendations, I’m your person.
You can reach out to me at saraswathi.nvr02@gmail.com
******
If you would like to volunteer to write for the BioCompute blog (which has 577 subscribers as of today) reach out here with a brief idea of what you would like to write about, and some writes samples from the past.
I can’t guarantee timelines for now (because we are doubling down on scaling up our data storage, and we don’t have a full time community person yet), but rest assured I will get back. Any messages received via Linkedin/email for this will not be considered.